news
Rules for Gene Silencing in Cancer Cells Identified
Primary tabs
Human cancers from breast and lung have a common pattern of genes vulnerable to silencing by DNA methylation, researchers at Emory University and the Georgia Institute of Technology have found. The results are published in the January issue of Cancer Research.
On their way to becoming tumors, cells have to somehow inactivate several "tumor suppressor" genes that usually prevent cancer formation. Methylation is a subtle punctuation-like modification of the DNA that marks genes for silencing, meaning that they are inactive and don't make RNA or proteins.
"We've developed a set of guidelines that allow us to predict which genes have an increased risk of silencing by DNA methylation," says senior author Paula Vertino, PhD, associate professor of radiation oncology at Emory University School of Medicine and Emory Winship Cancer Institute. "That vulnerability could make those genes good markers for diagnosis and risk assessment in patients."
The "signature" of DNA methylation found in cancer cells came from the team's previous work analyzing cell lines that artificially overproduce an enzyme which adds methylation markers to DNA. Vertino's team calls the signature PatMAn for "pattern-based methylation analysis."
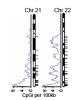
Dr. Eva Lee, associate professor and director of Center for Operations Research in Medicine and Healthcare of industrial and systems engineering at Georgia Tech developed the pattern recognition algorithm, feature selection, and predictive tools that pinpointed a small subset of DNA sequence signatures capable of classifying the methylation status.
In particular, Lee's approach identified PatMAn, which is based on seven "key words", 8-10 nucleotides long, that can predict which genes become methylated in breast and lung cancers in addition to artificial cell lines. Postdoctoral fellow Michael McCabe, PhD, took Lee's predictions and validated their status in the laboratory.
If the key words are in the DNA sequence near the promoter of the gene, it is more likely to be methylated. The promoter of a gene is the place where enzymes start making DNA into RNA.
Further analysis led to the team's realization that PatMAn overlaps with the pattern of DNA bound by a set of proteins known as the Polycomb complex in embryonic stem cells. Polycomb appears to keep genes that regulate early development turned off in embryonic stem cells.
The researchers combined PatMAn with the Polycomb binding pattern to generate SUPER-PatMAn, an improved version of PatMAn that could predict methylation-prone genes in cancers with more than 80 percent accuracy.
Vertino notes that the methylation pattern in cancer cells appears to echo Polycomb's binding in embryonic stem cells. Many of the genes affected play important roles in embryonic development.
"Many of the genes predicted by Lee's algorithms to be methylation-prone are developmental regulators," she says. "Our findings could support the idea that methylation-mediated silencing helps to lock the developmental state of tumor cells into being more stem cell-like."
Among cancer biologists, hypermethylation is now the most well characterized epigenetic change to occur in tumors. Lee's pattern recognition and classification tools offer the opportunity to classify the more than 29,000 known (but as yet unclassified) CpG islands in human chromosomes. This will provide an important resource for the identification of novel gene targets for further study as potential molecular markers that could have an impact on both cancer prevention and treatment. For aggressive cancers such as pancreatic cancer or some forms of incurable brain tumor, the ability to identify such sites offers potential new therapeutic interventions, leading to improved treatment.
The research was funded by the National Institutes of Health, the National Science Foundation, the American Cancer Society and the Georgia Cancer Coalition.
Reference: A multi-factorial signature of DNA sequence and Polycomb binding predicts aberrant CpG island methylation. McCabe, M.T., Lee, E.K, and Vertino, P.M, Cancer Research, 69(1): 282-291, 2009.
Status
- Workflow Status:Published
- Created By:Barbara Christopher
- Created:03/31/2009
- Modified By:Fletcher Moore
- Modified:10/07/2016
Keywords